My name is Mary Emeraghi, a PhD candidate at the University of Abomey-Calavi in Benin Republic. I am a Nigerian and a recipient of the GENES scholarship which I applied for and was graciously awarded in 2018. My research focuses on the maize crop. Maize (Zea mays L.) is an annual grass grown for its edible grains. It is the most important staple food on the African continent, grown and consumed by more than a hundred million smallholding families and also serves as a source for feed, fuel and fibre. My particular research interest is on conferring durability for maize streak virus (MSV) into locally adapted cultivars that may also bear a drought tolerant genetic background. MSV is considered as the most serious viral biotic constraint of maize that is endemic only to sub-Saharan Africa. Loss from MSV in an epidemic year can reach up to millions of dollars with maize yield losses of between 17 and 100%. I hope to navigate the various methods of improving for MSV resistance by firstly successfully introgressing some minor recovery resistance quantitative trait loci (QTLs) into collected maize germplasm grown in Benin and then advancing the developed hybrids up to the first backcross generation to recover the most of the local parent genome. Estimation of the combining abilities of the lines and testers used in the study will be done as well as heterosis to determine higher performing hybrids generated from the work. So far, the GENES scholarship has enabled me to gain sound understanding in hands-on molecular mechanisms needed for my laboratory work through my participation at the Marker-Assisted Training in a one-month training at BecA-ILRI hub in Nairobi, Kenya. I have also been privileged to carry out the phenotyping of collected donor and local maize genotypes for their resistance to the virus at the International Institute of Tropical Agriculture, Ibadan, Nigeria, all thanks to being under this scholarship. I am hopeful that the hybrids that will be generated by my work will help to position the maize production system of Benin Republic at a more climate-resilient vantage point and better secure and improve the livelihood of the more than 5 million smallholding families who depend on the crop.
About Mary Emeraghi
About Aboègnonhou Chaldia OdetteAGOSSOU
Aboègnonhou Chaldia OdetteAGOSSOU (MSc, Biotechnology and Plant Breeding)
Home Country: BeninSupervisor: Prof. Happiness O. Oselebe1 and Prof. Enoch Achigan-Dako2,
1Faculty of Agriculture & Natural Resources Management, Ebonyi State University, Nigeria
2Faculty of Agronomic Sciences, University of Abomey Calavi (UAC), Benin Republic
Professional Experience
Research Assistant at the Laboratory of Genetics, Horticulture and Seed Science (GBioS).
Scientific Publication
Agossou has published three articles in peer-reviewed Journals.
Crop of interest
My research focuses on Macrotylomageocarpum, an orphan grain crop belonging to the Fabaceae family. This crop is a valuable one with a kg of the grains reaching up to 8 USD. It is a well consumed grain in Benin, where it is used as prestigious events food. The crop is also used in folk medicine by few communities in West Africa, included Nigeria, Togo, Ghana and Burkina Faso. Recently the crop has benefited from an increasing interest in breeding areas with focus on the seed yield and the nutritional quality improvement.
Research topic
Yield improvement and protein variation analysis in kersting’s groundnut [Macrotylomageocarpum (Harms) Maréchal and Baudet] in West Africa.
Summary of proposal
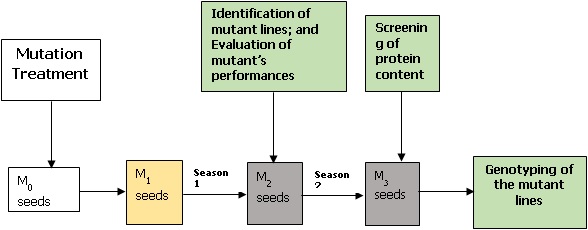
Most Africa countries have been facing two challenges, feeding their increasing population and overcoming the impact of climate variability. One way to contribute to increasing the food security in tropical areas, especially in Africa under the new climate change is to increase interest for breeding of some orphan crop that have the potential to easily contribute to the diets diversification and the nutritional status of local population improvement. Though legumes are an important commodity group owing to their multi-functionalities, many of the crops are still under-researched, including the kersting groundnut. Our project intends to develop improved seed yield of kersting groundnut by inducing mutation within the species; analyzing the variation in protein content within the crop and identifying SNP markers linked to this trait through genome wide association study. Our development objective is to provide farmers with high yielding and protein-dense seed.A graphical abstract of my research methods is shown below.

Personal Comment
I am grateful to the Project Team who worked hard to bring this kind of nice project sponspored by European Union which gave me this kind of opportunity. I’m excited to be one of the scholars of this amazing program which will improve my skill and capacity in molecular breeding.
Fentanesh Chekole KASSIE (MSc, Plant Biotechnology)
Home Country: NigeriaSupervisor: Dr. Mesfin Kebede1,Prof. Joseph Martin Bell,Dr. Hermine Bille and Dr. Joel Romaric Nguepjop 2
1Jimma University, Ethiopia
2University of YaoundeI, Cameroon.
Professional
Lecturer, Wolaita Sodo University,Ethiopia.
Crop of Interest
Peanut, Arachishypogaea L., is an important grain legume providing edible oil and protein for human nutrition. Peanut is an important oil, food and feed crop of the world. The kernels are rich in fats and protein, and 100 g of kernels provide 567 kcal of energy and 8.5 g of dietary fibre. Consumption of peanuts can reduce risk of inflammation, diabetes, cancer. Peanuts have a variety of industrial end uses. Paint, varnish, lubricating oil, leather dressings, furniture polish, insecticides and nitro-glycerine are made from peanut oil.
Research Topic
Genome wide QTL mapping of seed quality and yield-related traits in a peanut Advance Backross population
Summary of proposal
Peanut, Arachis hypogaea L, is an important grain legume providing edible oil and protein for human nutrition. The limited genetic diversity in cultivated gene pool has greatly hampered the application of molecular tools for breeding. However, there are wild relatives with a high level of genetic diversity. They are an important source for broadening the genetic base of cultigen and represent a great reservoir of alleles that can be used for the improvement of simple and more complex traits. To minimize the confounding effects of deleterious traits, advanced backcross (AB) quantitative trait loci (QTL) analysis was developed. An AB-QTL analysis was applied in peanuts and several QTLs with favourable alleles from the wild donor parent were identified. However, there is little effort to improve peanut quality traits. The ultimate goal of this work is to identify QTL/genes involved in the inheritance of seed quality and yield related characters using an AB-QTL population. A set of 27 cultivated varieties,10 wild relatives and 133 AB-QTL lines will be grown in Cameroon.A total of 38 traits related to flowering, plant and leaf architecture, pod and seed morphology, biochemistry (seed quality traits) and yield components,Genomic DNA will be extracted and genotyped, with Single Nucleotide Polymorphism.
Personal Comment about the GENEs Opportunity
The GENEs project gave very good chance to me to study plant genomics,which is very good for me because I have MSc degree related in plant genomics. In addition to geting knowledge about plant genomics GENEs project opened the door to share different culture and technology.
Mahaman Mourtala Issa Zakari (Mphil, Plant Breeding)
Home Country: NigerSupervisor: Prof. H. O. Oselebe1 and Prof. Barrage Moussa 2
1Faculty of Agriculture & Natural Resources Management, Ebonyi State University, Nigeria
2Faculty of agricultural Sciences, Abdoul Moumouni University of Niamey, Niger
Professional Experience
Research Engineer (National Institute of Agricultural Research of Niger)
Assistant Manager, plant genetic resources National Gene bank of Niger,
National focal point of International Treaty for Plant Genetic Resources for Food and Agriculture (ITPGRFA/FAO), Niger
Scientific Publication
Issahas published a paper in Academic Journal of Plant Breeding and Crop Science (JPBCS).
Crop of Interest
Sweetpotato [Ipomoea batatas (L.) Lam] is a perennial plant cultivated as annual crop andbelongs to morning glory family Convolvulaceae. It is an important tuber crop grown in more than 110 countries in the word. It is an important food, feed and vegetable crop in most tropical developing countries. The crop is ranked fifth economically after rice, wheat, maize, and cassava. The greens of the crop are edible and provide an important source of food in Africa, especially in Guinea, Sierra Leone and Liberia, as well as in East Asia. In East Africa, thousands of villages depend on sweetpotato for food security and it is called [cilera abana[, "protector of the children". In third world countries, sweet potato is processed into starch, noodles, candy, desserts, and flour, which allow the farm household to extend the availability of the crop. In some countries, the storage roots are processed to produce starch and fermented to make alcohol. In China, sweetpotato starch production has become an important cottage industry. In Japan, 20% of sweetpotato produced are used for starch production. The juice of red sweet potato is combined with lime juice in South America to make a dye for cloth; and every shade from pink through purple to black can be obtained.The sweetpotato forage shows dry matter contents of 12 - 17% and dry matter digestibility of greater than 70%.It provides high calorie at 152 MJ ha-1 day-1 compared to cassava, wheat, rice and maize with respectively 121, 135, 151, and 159 MJ Ha-1 day-1 calories. The storage roots are rich in carbohydrate and its leaves are rich in proteins. Orange and yellow fleshed varieties are rich in Beta-carotene, a precursor of vitamin A, while purple-fleshed varieties contain anthocyanin, which is a powerful anti-oxidant and the white-fleshed is rich in dry mater. These sweet potato are being used in a number of African countries to combat the widespread Vitamin A deficiency that results in blindness and even death of 250,000-500,000 African children a year.
Summary of proposal
More than 70% of potential yield loses in agriculture worldwide is attributed to adverse environmental factors of which water scarcity represents the most severe constraint. Drought is therefore often a major environmental constraint for sweetpotato production in areas where it is grown under rain fed condition.Development of improved, drought tolerant sweetpotato varieties will increase sweetpotato yields especially in arid and semi-arid lands where seasonal drought is a significant problem. In Niger and Nigeria, local varieties are the most used and fail to meet the demand of end-users for different utilization purposes. The main objective of the study is to develop higher yielding drought tolerant sweetpotato varieties that are acceptable by farmers and consumers in West Africa. Simple Sequence Repeat markers will be used to assess the genetic diversity and to select suitable parents. Accelerated Breeding Scheme will be the breeding method which can allow cultivarrelease in 3-4 years that conventional breeding cannot (7-8 years). Fifteen parents will be crossed using diallel mating design Model 1 Method 1to develop the hybrids. The F1 progenies will be evaluated under drought stress at constrained number of location (following these steps: observational trial, preliminary yield trial and advanced yield trial).After evaluation, the best hybrids will be selected for multilocational and on-farm trials. The new hybrid varieties will then be registered in the regional catalogues of cultivated species and varieties (ECOWAS) and released to farmers. This can allow farmers to increase their production to meet their demand, earn money and process to industrial use. The Hybrid types that will be developed would provide Beta-carotene, contain anthocyanin and high dry mater.
















































